Epigenetics systems including DNA methylation, histone modifications and nucleosome positioning comprise the non-genetic memory of a cell. These mechanisms are reversible, heritable and responsive to developmental, behavioral and environmental cues. Variations in epigenetic profiles are critical determinants of cell lineage commitment, phenotypic variability and susceptibility to disease. Epigenetic modifications function to regulate gene expression by causing changes in chromatin structure that alter the accessibility of cis-acting DNA elements to trans-acting factors. Every gene has underlying epigenetic components that mechanistically contribute to its regulation. Therefore, researchers studying gene regulation will eventually find themselves exploring the epigenetics of gene regulation.
One of the more established hallmarks of epigenetic regulation is post-translational modification (PTM) of histones. Histones are responsible for packaging DNA into nucleosomes, the basic structural unit of chromatin. They are subject to a variety of PTMs, including phosphorylation, acetylation, and methylation at specific amino acid residues on the histone tails that protrude from the core nucleosome. These modifications lead to steric changes in chromatin structure that regulate various cellular processes such as transcription, replication and DNA repair. The combinatorial pattern of histone PTMs and histone variants function as a ‘histone code’ that instructs remodeling and binding events at specific chromosomal regions. In doing so, PTMs alter either the accessibility of underlying genes to the transcriptional machinery or the recruitment of effector proteins that either recognize (“reader”), deposit (“writer”) or remove (“eraser”) these histone marks.
Epigenetics and Human Disease
Histone modifications have important implications for human health and disease. Mounting evidence exists to link abnormalities in histone PTMs and histone modifying enzymes to human pathologies, including developmental, autoimmune, neurological, inflammatory and neoplastic disorders. The importance of studying histone modifications is evident in various research areas. Histone modifications are frequently targeted in biomarker discovery studies as predictors of cancer prognosis, as markers of active (H3K4me3) or repressed (H3K9me3 and H3K27me3) chromatin, and as a significant component of cell signaling responses that lead to changes in gene expression.
Relative to DNA methylation, far less is known about the biological significance of histone PTMs, mostly due to limitations in available technologies. Much of the published work on histone PTMs is based on evidence provided by traditional methods such as Western blot, immunohistochemistry and genome-wide mapping. However, these methods are time-consuming, lack high throughput capabilities and can be quite costly when interrogating more than one histone PTM target. To address some of these limitations, Active Motif recently partnered with Luminex to develop the first multiplex epigenetic assay for the study of histone PTMs using either the Luminex® 200™ or MAGPIX® instruments.
Multiplex Epigenetic Assay
The Histone PTM Multiplex Kit is a highly sensitive assay that enables high throughput processing of histones using low sample amounts (nanogram quantities) to study multiple histone modifications within a single well. The assay format also offers the ability to normalize Histone PTM data against total histone H3 levels when comparing different samples or treatment conditions.
The Histone PTM Multiplex Assay works as a solution-based sandwich ELISA to interrogate the levels of histone modifications within acid extracted cell lysates or purified histones. Histone PTM-specific antibodies have been conjugated to unique Luminex® xMAP microspheres and are used to bind the N-terminal histone modifications within the sample. A biotinylated antibody specific for the C-terminus of Histone H3 is then added to bind the captured histone and form a ‘sandwich’. Next, streptavidin-phycoerythrin is introduced into the reaction to bind the biotinylated reporter antibody. The MFI readout is proportional to the amount of captured histone PTM. Since each histone PTM is associated with a unique bead set, this system allows for the addition of multiple Histone Antibody-conjugated Beads to each sample for multiplexing.
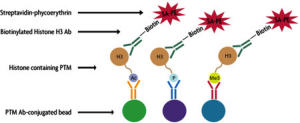
Histone Antibody-conjugated Bead Sets include targets for: Total Histone H3, H3K4me3, H3K27me2, H3K27me3, H3K9me1, H3K9me2, H3K9me3, H3S10ph, H3K9ac and H3 pan-acetyl marks.
To learn more about Active Motif’s Histone PTM Multiplex Assay, please visit http://www.activemotif.com/search?terms=Luminex+Services.