As new drugs and combination therapies are developed for the treatment of metastatic colorectal cancer, the effectiveness of certain therapeutic compounds is dependent on the genetic characteristics of target tumors. This is exemplified by studies showing different levels of response to treatment with therapeutic anti–epidermal growth factor receptor (anti-EGFR) monoclonal antibodies such as cetuximab and panitumumab. A number of studies have identified activating mutations in KRAS and several other EGFR signaling pathway genes that are associated with tumor resistance to treatment with anti-EGFR antibodies.1-4
As a result of studies like these, the American Society of Clinical Oncology (ASCO) published a provisional clinical opinion outlining the need for genetic testing of KRAS mutations to determine which metastatic colorectal cancer patients could benefit from treatments that include anti-EGFR therapeutics.5 This opinion specifically addressed the need for identifying activating mutations in KRAS codons 12 and 13 associated with anti-EGFR resistance. It also mentioned that activating mutations occur in KRAS codons 61 and 146 as well as in BRAF, PI3K, and PTEN. The opinion mentioned these additional mutations; however, it did not provide guidance on screening details for the additional KRAS, BRAF, PI3K, and PTEN mutations or treatment strategies for different genotypes.
In addition, the ASCO opinion of 2009 did not address the differences in sensitivity or specificity of different assays for tumor genotyping analysis. It encouraged oncologists to consult with their clinical laboratory directors about what assays to use. As a result of these recommendations, a number of papers have been published comparing different assay technologies for tumor genotyping. Some of these assay technologies include direct sequencing, real time PCR (RT-PCR), RFLP analysis, High-resolution Melting Analysis, and a PCR-based multiplex Luminex® assay.
While direct sequencing has been the standard assay approach, it is a high cost per sample platform and is not amenable to running a large number of samples in a short period of time. Some of the least expensive approaches include standard RT-PCR and RFLP analysis approaches like those described in Lanthaler et al.6 When these RT-PCR and RFLP approaches were tested against each other, concordance of the number of subjects with a KRAS codon G13D mutation ranged from 77.4% to 93.5%. The improved concordance was achieved in part by reselection of tumor material that contained more tumor cells indicating a lack of sensitivity for these assays. While these approaches are fast and inexpensive, more accuracy with fewer false positive and negative calls of mutation status is required.
Several groups have compared direct sequencing results with that of High-Resolution Melting Analysis (HRMA) of mutations in KRAS and several other genes associated with anti-EGFR resistance.7,8 HRMA assays rely on the comparison of differences in the melt curves for PCR products from mutants with that of wild type controls. In Akiyohi et al., the authors report a concordance of 94.8% between the two assays for several KRAS mutations.7 The lack of 100% concordance required confirmation of discrepant HRMA calls by direct sequencing. While their analysis indicated that HRMA is fast and inexpensive, a few characteristics of the assays may contribute to its not being as accurate for some mutations. For example, the authors noted that it was difficult for HRMA to distinguish between two mutations adjacent to each other when both are present on KRAS codons 12 and 13.
While some of these methods are rapid and inexpensive, weaknesses in sensitivity and specificity raise concerns about their utility if not complemented with other assays to verify tumor genotype. In addition, these assays are also single plex, which require a separate reaction for each mutation being analyzed from a sample. This requires using more tumor sample material from tumor biopsies. Since the accuracy of these assays is also dependent on isolating DNA from tumor sections that contain different amounts of tumor cells, an assay that can use less sample material from tumor cell rich areas of a biopsy and provide accurate information on multiple genes is needed. Such an assay was tested by Bando et al.9 These authors developed a Luminex-based multiplex assay that can analyze 36 mutations in KRAS, BRAF, NRAS, and PIK3CA in one reaction, and compared it to direct sequencing (Table 1).
Table 1. List of KRAS, BRAF, NRAS, and PIK3CA analyzed with the Luminex assay.
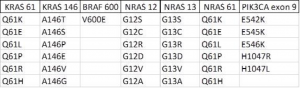
Analysis of these 36 mutations was performed on 78 surgically resected specimens and five biopsy specimens from 82 subjects. The tumor genotypes of these mutations obtained with the Luminex assays were in 100% agreement with that obtained by direct sequencing. The accuracy of the Luminex assay is in part due to the simple PCR chemistry and capture of specific mutation amplicons on Luminex beads. Additionally, each PCR reaction needs only 50 ng of genomic DNA to detect all 36 mutations in the same reaction, which decreases the amount of sample needed, the cost, and the time it takes to analyze multiple samples.
This ability to rapidly and accurately analyze multiple genes in one sample allowed the authors to make additional correlations between the occurrence of certain mutation combinations in the tumors and the correlation of these combinations to survival rates. It also demonstrated the platform’s ability to generate accurate tumor genotype information for multiple genes with less sample than other methods. In a clinical situation where the amount of a tumor sample may be limiting, the ability to detect mutations in multiple genes of clinical importance is of significant benefit to making important treatment decisions. With the Luminex platform’s ability to analyze up to 500 markers in one reaction, it is possible to add more markers that may be needed for determining the susceptibility or resistance of additional tumor genotypes to certain compounds in the future.
Interested in developing your own assays? The free xMAP® Cookbook contains detailed methods and protocols to help you develop assays on the xMAP platform.
References
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res 2007;67(6):2643-2648.
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26(35):5705-5712.
- Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 2009;101(4):715-721.
- Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69(5):1851-1857.
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27(12):2091-2096.
- Lanthaler A, Spizzo G, Mitterer M, Mian C, Mazzoleni G. Interlaboratory comparison of K-ras testing by real-time PCR and RFLP in colorectal cancer samples. Diagnostic Molecular Pathology 2011;20(2):90-93.
- Akiyoshi K, Yamada Y, Honma Y, Iwasa S, Kato K, Hamaguchi T, Shimada Y, Taniguchi H, Furuta K. KRAS mutations in patients with colorectal cancer as detected by high-resolution melting analysis and direct sequencing. Anticancer Res 2-13;33(5):2129-2134.
- Guedes JG, Veiga I, Rocha P, Pinto P, Pinto C, Pinheiro M, Peixoto A, Fragoso M, Raimundo A, Ferreira P, Machado M, Sousa N, Lopes P, Araujo A, Macedo J, Alves F, Coutinho C, Henrique R, Santos LL, Teixeira MR. High resolution melting analysis of KRAS, BRAF, and PIK3CA in KRAS exon 2 wild-type metastatic colorectal cancer. BMC Cancer 2013;13:169.
- Bando H, Yoshino T, Shinozaki E, Nishina T, Yamazaki K, Yamaguchi K, Yuki S, Kajiura S, Fujii S, Yamanaka T, Tsuchihara K, Ohtsu A. Simultaneous identification of 36 mutations in KRAS codons 61and 146, BRAF, NRAS, and PIK3CA in a single reaction by multiplex assay kit.” BMC Cancer 2013;13(1):405.